
NALgen
NALgen (Neutral and Adaptive Landscape Genetics) is a method for modeling
the process of gene flow and predicting the resulting pattern of (neutral and
adaptive) genetic variation across (continuous) landscapes.

NALgen:
Analysis Steps
To evaluate the NALgen method of modeling neutral and adaptive genetic variation across
continuous landscapes:
Generated several landscapes, and then simulated genotypes based on the generated landscapes
Used multivariate analyses
to infer population structure in the simulated data
to find covariance between genetic and environmental data.
Modeled neutral and adaptive genetic variation across continuous landscapes
Information about population structure used to create response variable
Transformed environmental data (based on genetic-environmental covariance) used as
predictors
Evaluated performance of model (using subsets of 20 randomly-distributed populations)
Compared against original landscape used to simulate genotypes
Compared against inferred map of ancestry based on genotypes simulated for each cell
(all 884 populations)
Landscape
Genetic
Simulations
Generating simple (low-complexity) landscapes and simulating genotypes
landscapeRpackage: simulating simple landscapes (barrier and two
different values of carrying capacity/habitat suitability): landscape 1 has
low permeability cells (carrying capacity = 5) between habitable patches
(carrying capacity = 100), while landscape 2 has high permeability cells
(carrying capacity = 25), in addition to barriers (carrying capacity = 0)
popRangepackage: simulating 50selectedand 500neutralSNPs based
on thelandscapeR-generated landscape.
Generating high-complexity landscapes and simulating genotypes
virtualspeciespackage: landscape generation based on Bioclim data
Collinearity removed from climatic variables,
PCA of remaining variables performed,
Ranges of PCA values specified (for three metapopulations).
popRangepackage: simulating 50selectedand 500neutralSNPs based on
thevirtualspecies-generated landscape.
Simulated genotypes (50 selected and 500 neutral biallelic loci (SNPs), separately and combined): two migration coefficients
(m=0.1 and 0.5) x two levels of landscape complexity (low and high) x two levels of landscape permeability (low and high)
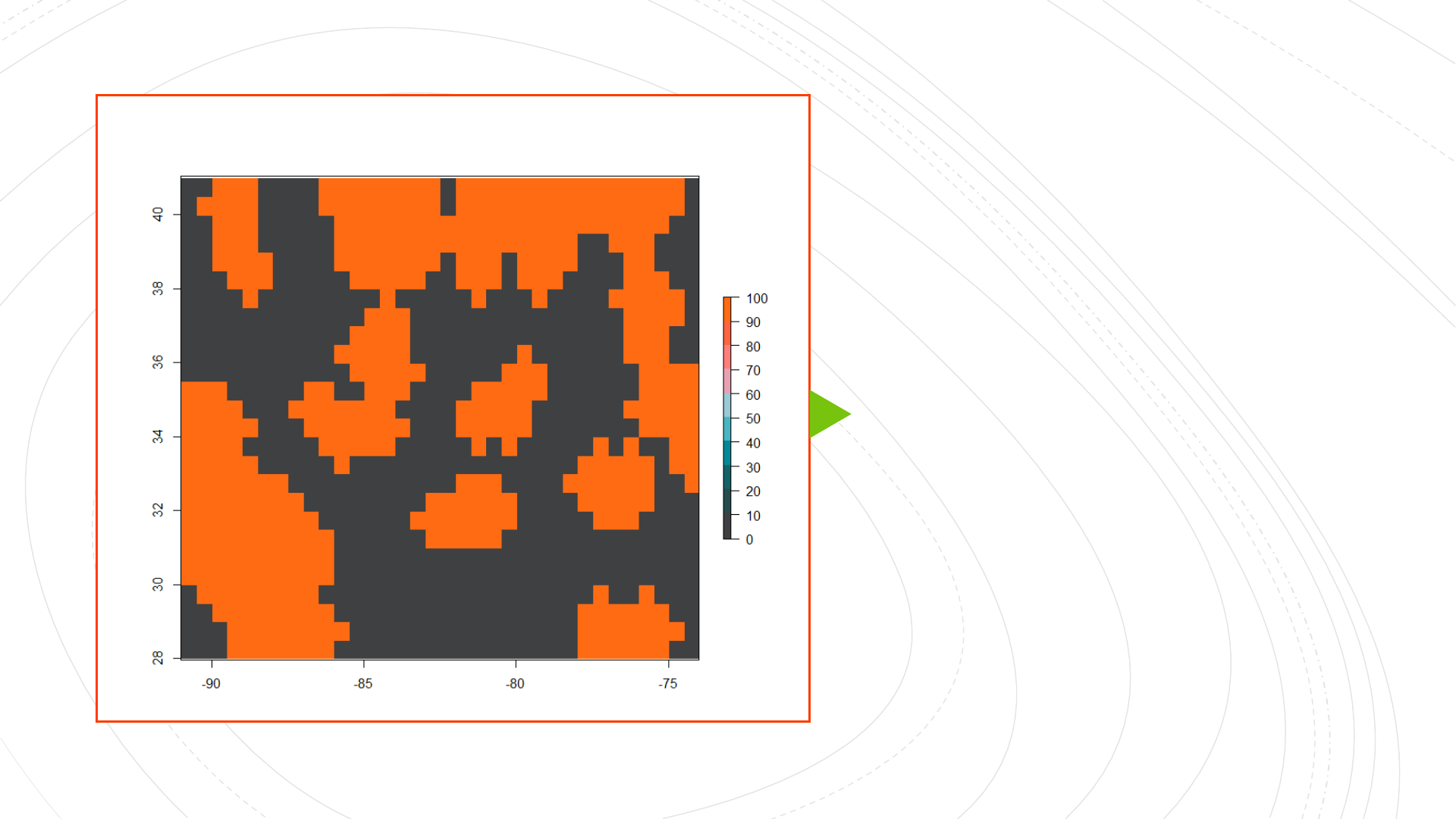
Low-Complexity
Low-Permeability
(LCLP) landscape
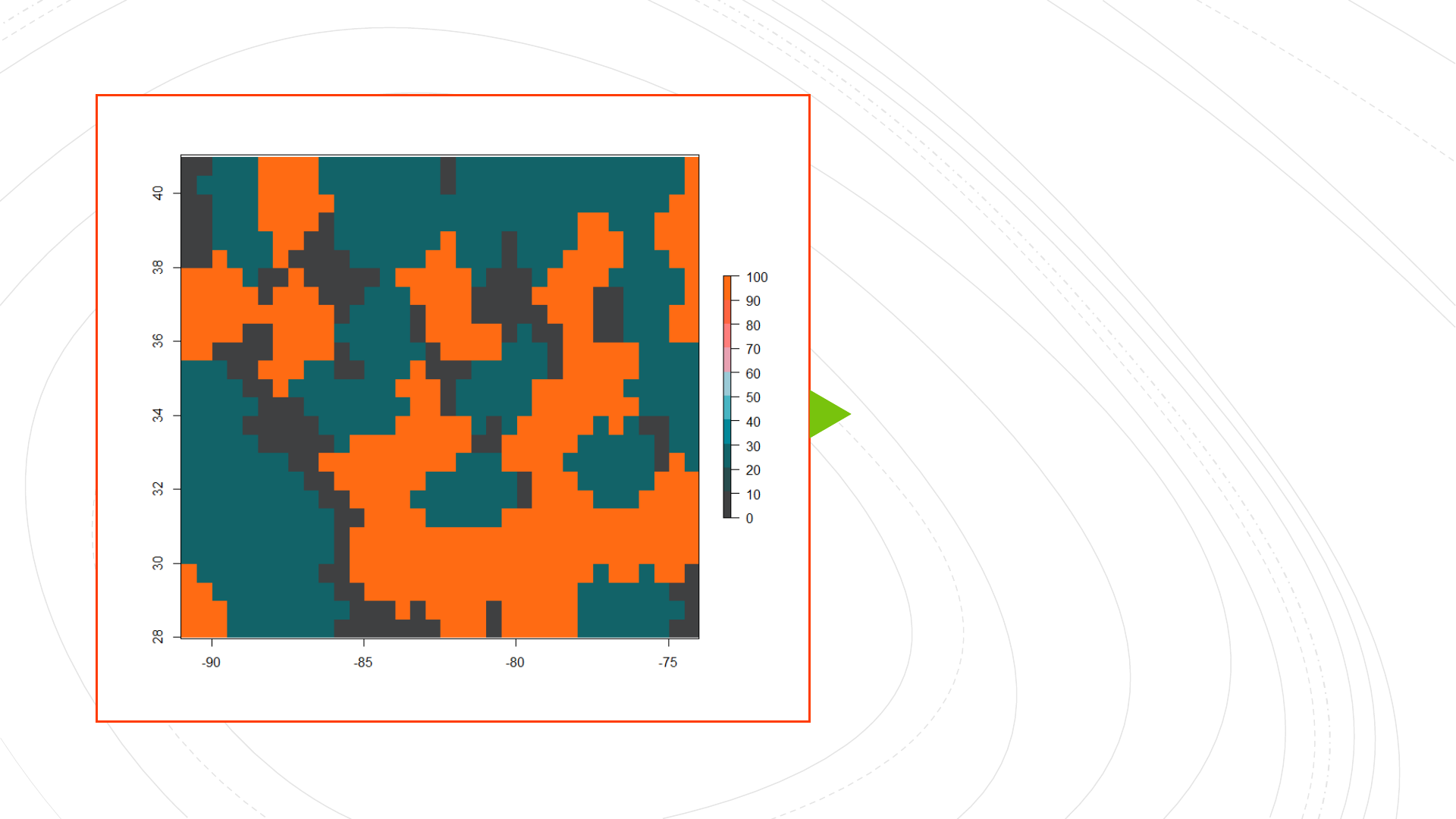
Low-Complexity
High-Permeability
(LCHP) landscape
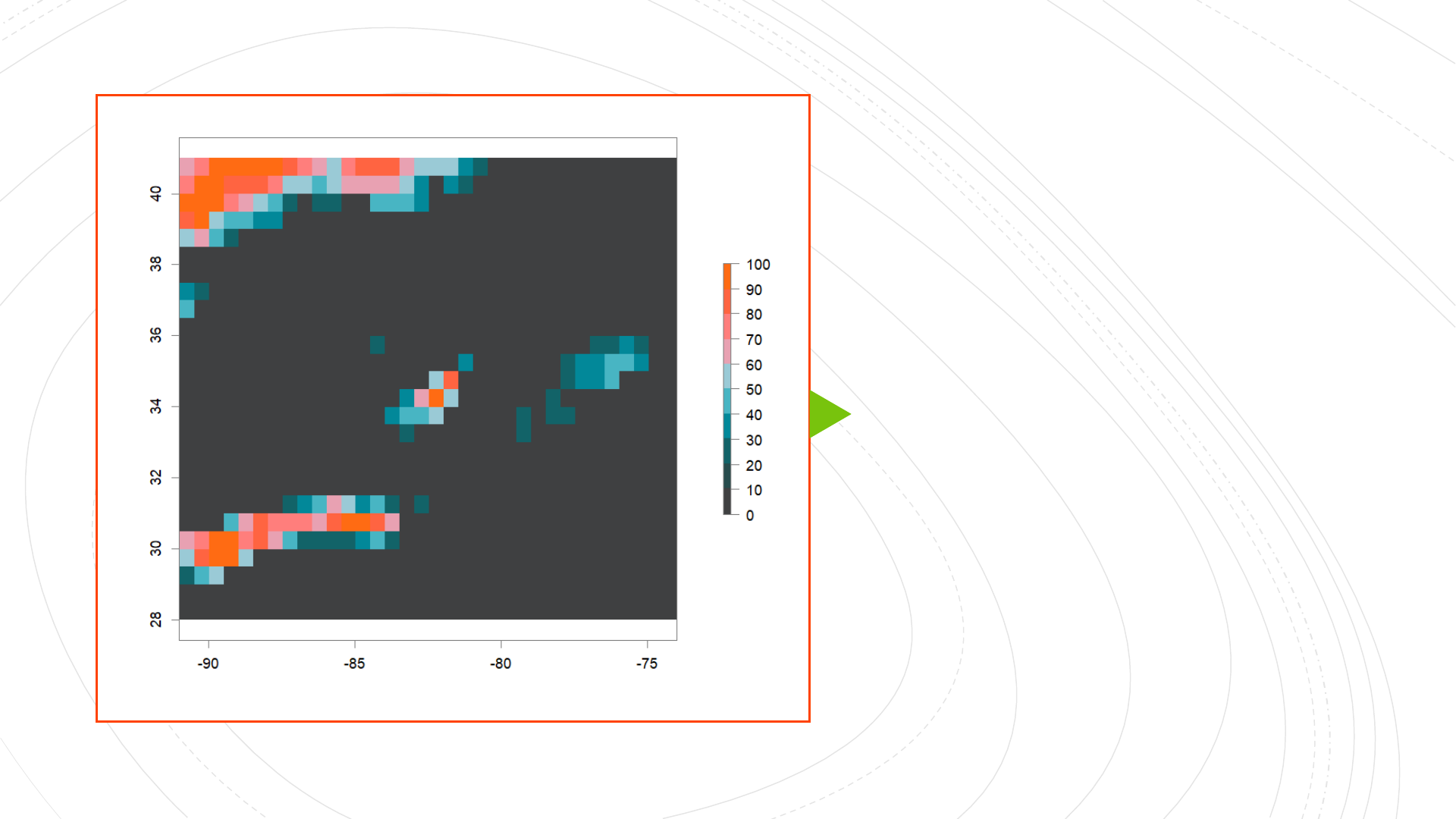
High-Complexity
Low-Permeability
(HCLP) landscape
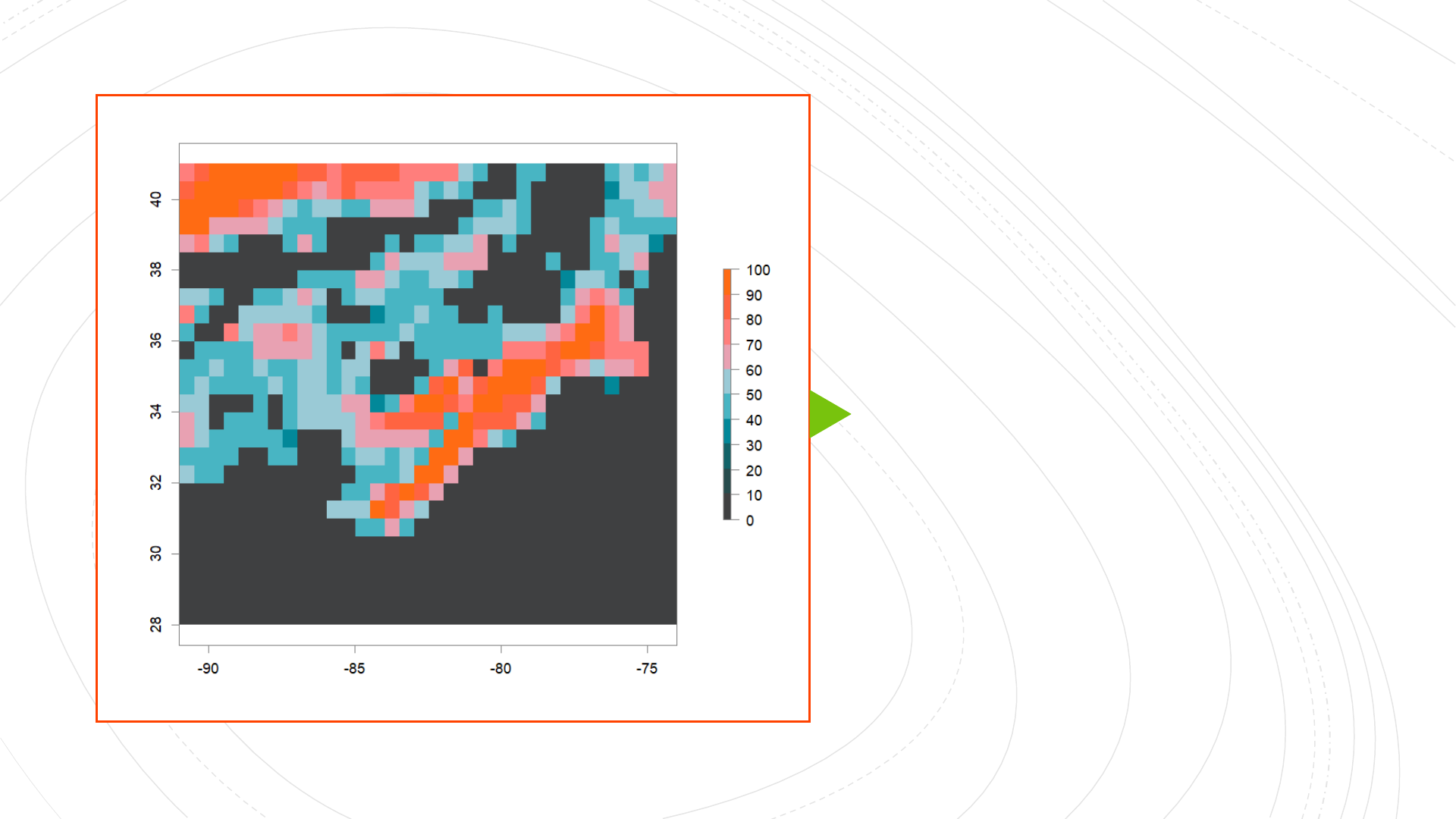
High-Complexity
High-Permeability
(HCHP) landscape

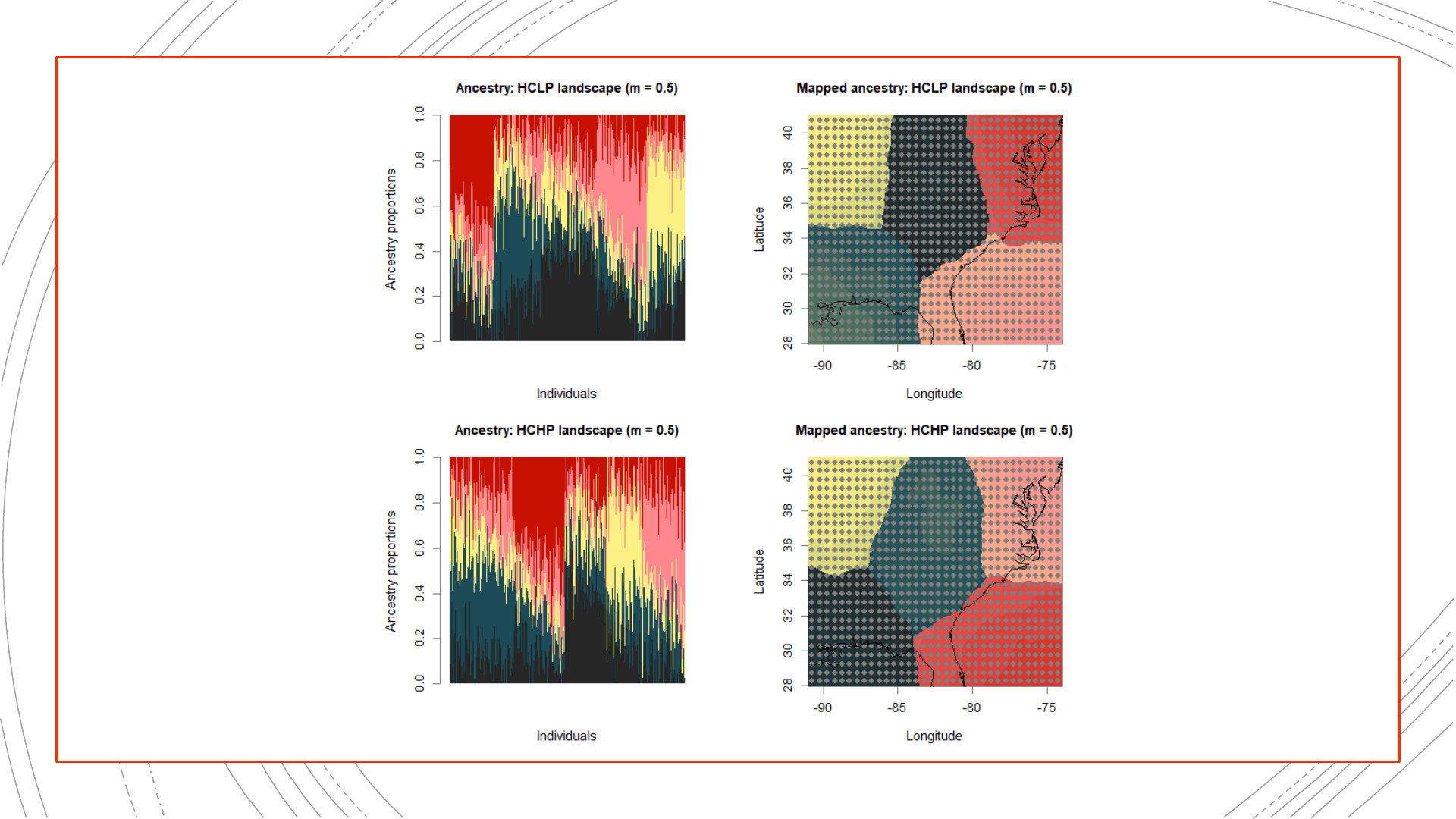
Setup of Evaluation Step:
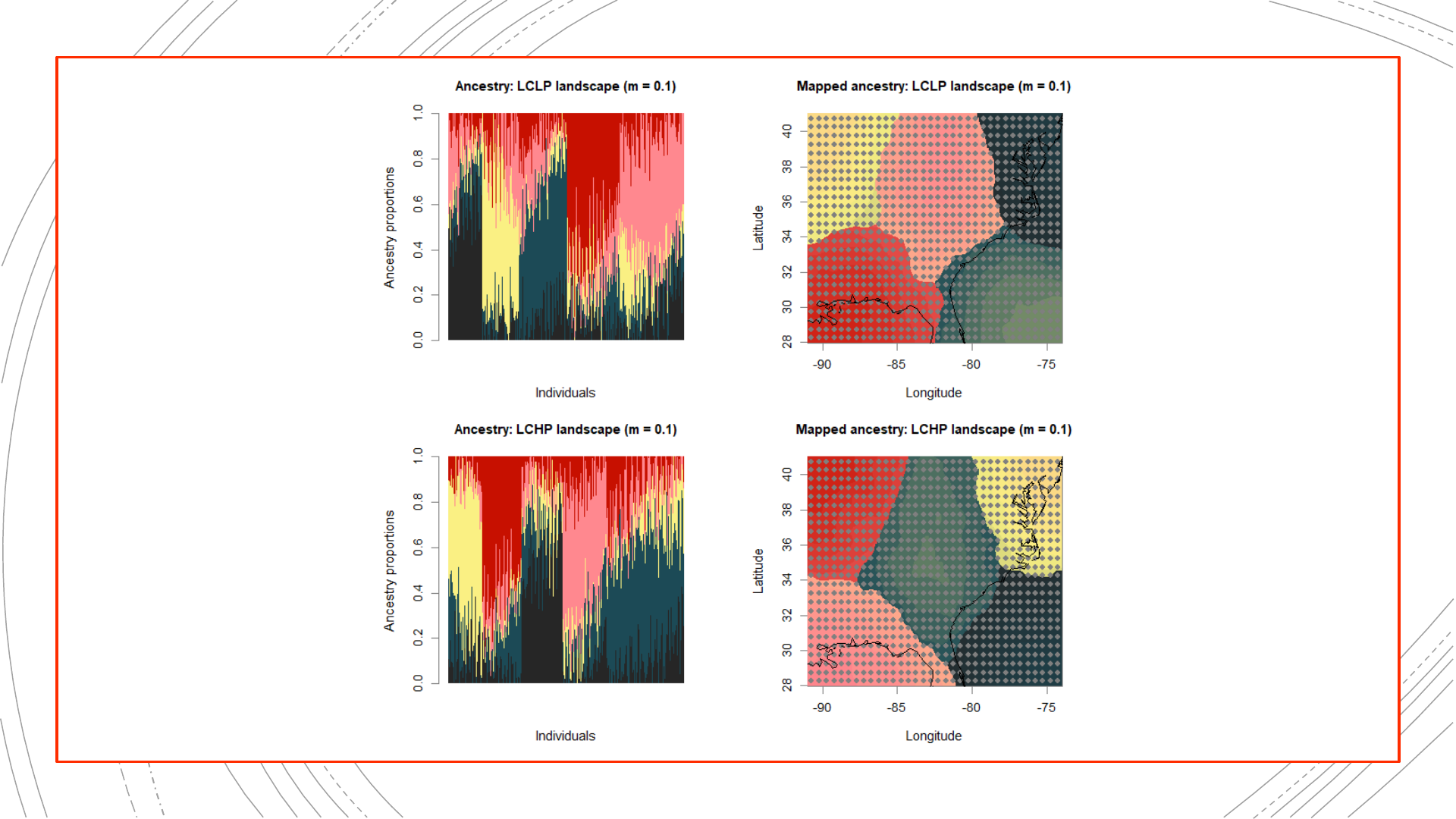
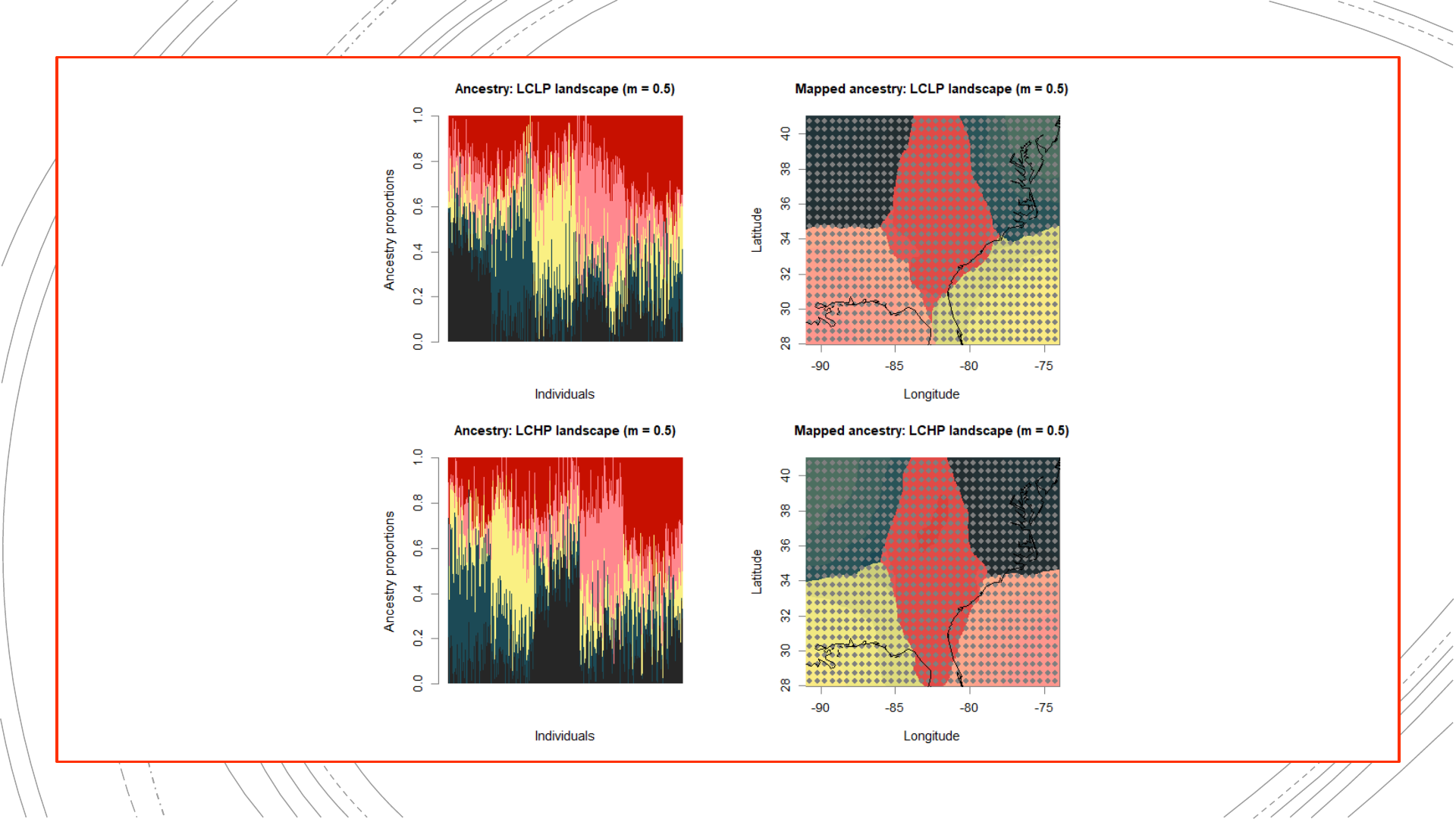
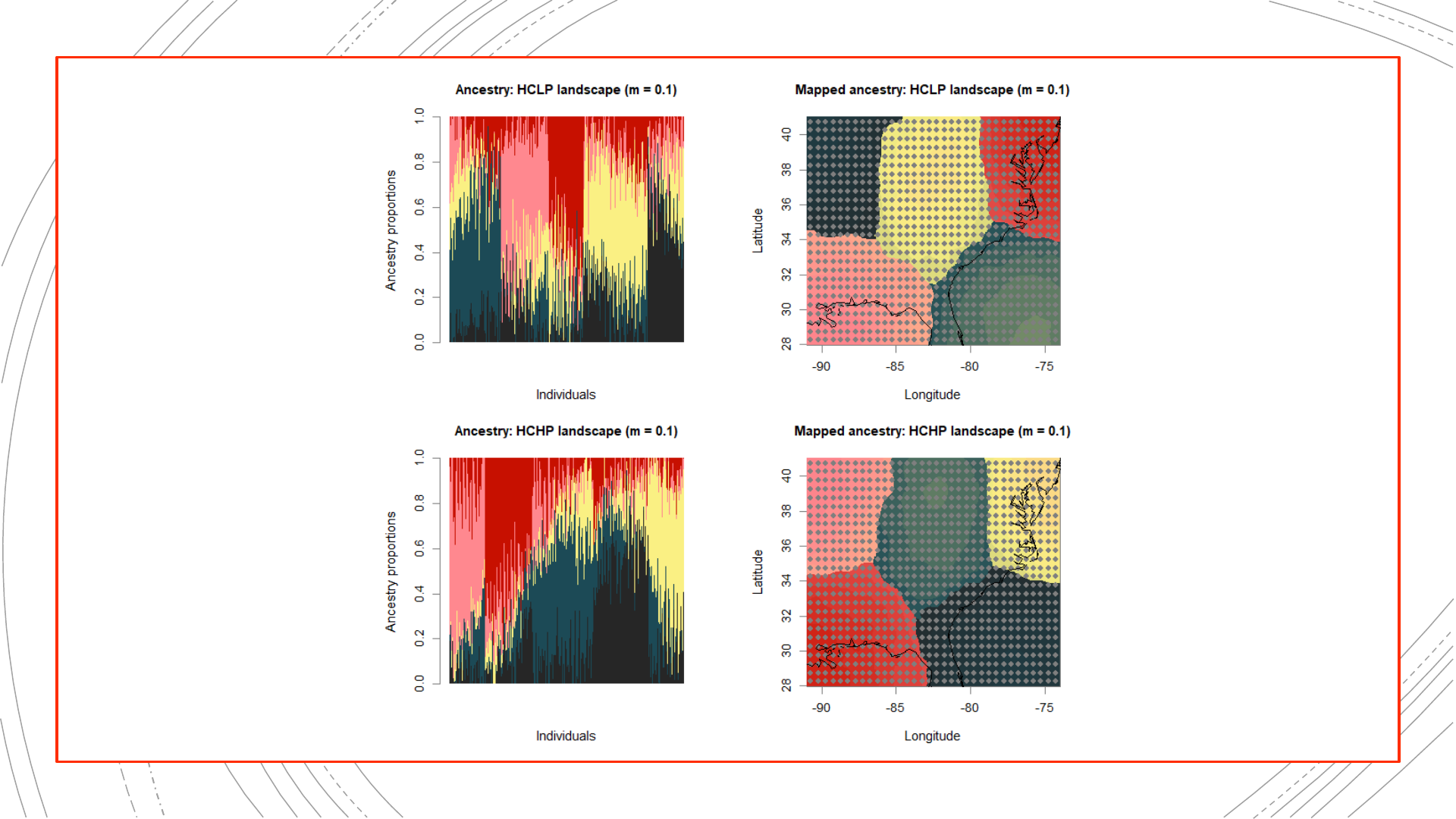
884-population reference
The original landscapes used to
simulate the genetic data, as well
as population structure results
obtained from these complete
genetic datasets, will be used in
evaluating the NALgen method.
Presented hereinafter are the
reference results (mapped
ancestry coefficients) that NALgen
modeling based on 20 randomly-
distributed populations will be
compared against.

NALgen Method
To determine the maximum genetic-
environmental covariance, the raw data is
first transformed: these transformed
environmental scores form the predictors
in NALgen modeling
The response variables are measures of
genetic variation (neutral and adaptive):
these are newly-developed metrics